class: center, middle  #### DSTA Extra Materials ### Evaluating Classification performance Extra Slides and codes courtesy of [Andreas C. Müller, NYU](https://github.com/amueller/) --- class: centre,middle ### Supervised Binary Classification __Istance:__ - a collection __X__ - its classification __y__ (assume a classification sys. {0,1} or {-1, +1}) __Solution:__ a classifier function (here called *model*) __Measure:__ misclassification wrt. __y__ .center[  ] --- ### Confusion matrix .center[  ] ??? Diagonal divided by everything. --- #### Example: evaluate the *LogisticRegression* classifer .smaller[ ```python from sklearn.datasets import load_breast_cancer from sklearn.linear_model import LogisticRegression data = load_breast_cancer() ``` ] --- #### Example: evaluate the *LogisticRegression* classifer .smaller[ ```python from sklearn.datasets import load_breast_cancer from sklearn.linear_model import LogisticRegression data = load_breast_cancer() ``` ] .smaller[ ```python X_train, X_test, y_train, y_test = train_test_split( data.data, data.target, stratify=data.target, random_state=0) lr = LogisticRegression().fit(X_train, y_train) y_pred = lr.predict(X_test) ``` ] --- #### Example: evaluate the *LogisticRegression* classifer .smaller[ ```python from sklearn.datasets import load_breast_cancer from sklearn.linear_model import LogisticRegression data = load_breast_cancer() X_train, X_test, y_train, y_test = train_test_split( data.data, data.target, stratify=data.target, random_state=0) lr = LogisticRegression().fit(X_train, y_train) y_pred = lr.predict(X_test) ``` ] .smaller[ ```python from sklearn.metrics import confusion_matrix print(confusion_matrix(y_test, y_pred)) print(lr.score(X_test, y_test)) ``` ]  --- #### Problems with Accuracy Data with 90% positives: .smaller[ ```python from sklearn.metrics import accuracy_score for y_pred in [y_pred_1, y_pred_2, y_pred_3]: print(accuracy_score(y_true, y_pred)) ``` ``` 0.9 0.9 0.9 ``` ] .center[  ] ??? - Imbalanced classes lead to hard-to-interpret accuracy. --- class:split-40 ### Precision, Recall, f-score .left-column[ `$$ \large\text{Precision} = \frac{\text{TP}}{\text{TP}+\text{FP}}$$`<br /> `$$\large\text{Recall} = \frac{\text{TP}}{\text{TP}+\text{FN}}$$`<br /> `$$\large\text{F_1} = 2 \frac{\text{precision} \cdot\text{recall}}{\text{precision}+\text{recall}}$$`] .right-column[ <br /> Positive Predicted Value (PPV) <br /> <br /> <br /> Sensitivity, coverage, true positive rate. <br /> <br /> <br /> Harmonic mean of precision and recall ] ??? All depend on definition of positive and negative. --- #### F-score `$$\large F_1 = 2 \frac{\text{precision} \cdot\text{recall}}{\text{precision}+\text{recall}} = \frac{2}{\frac{1}{\text{precision}}+\frac{1}{\text{recall}}}$$` Prize on higher recall: `$F_{2}$` Prize on higher precision: `$F_{0.5}$` --- class: center, spacious #### The Zoo  https://en.wikipedia.org/wiki/Precision_and_recall ??? --- class:split-40 .right-column[  <br />  <br /> 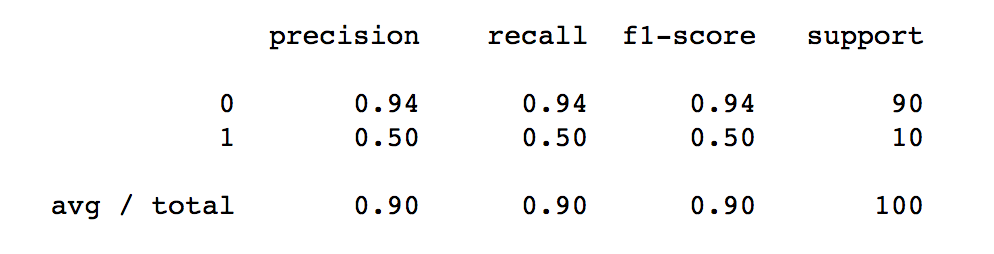 ] .left-column[ 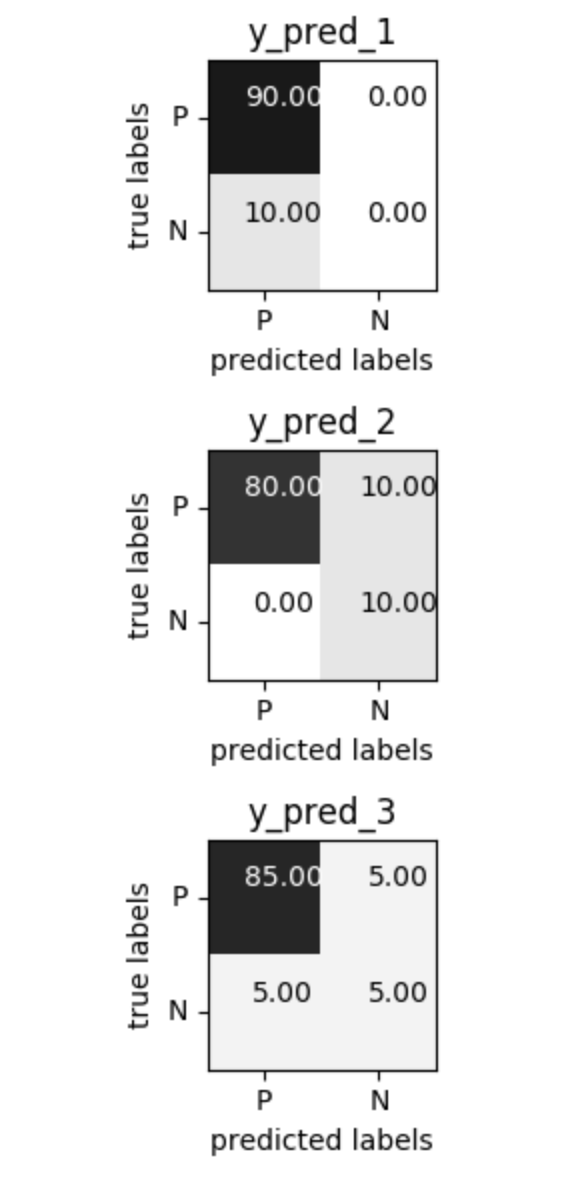 ] ??? --- class:spacious #### The imporante of experiment design - What do I want? What do I care about? - Can I assign costs to the confusion matrix? - What guarantees do we want to give? (the following examples are for `data = load_breast_cancer()`) ??? (precision, recall, or something else) (i.e. a false positive costs me 10 dollars; a false negative, 100 dollars) --- #### Setting Thresholds .tiny-code[ ```python X_train, X_test, y_train, y_test = train_test_split( data.data, data.target, stratify=data.target, random_state=0) lr = LogisticRegression().fit(X_train, y_train) y_pred = lr.predict(X_test) print(classification_report(y_test, y_pred)) ``` ``` precision recall f1-score support 0 0.91 0.92 0.92 53 1 0.96 0.94 0.95 90 avg/total 0.94 0.94 0.94 143 ``` ```python y_pred = lr.predict_proba(X_test)[:, 1] > .85 # predict 1 only when Pr>.85 print(classification_report(y_test, y_pred)) ``` ``` precision recall f1-score support 0 0.84 1.00 0.91 53 1 1.00 0.89 0.94 90 avg/total 0.94 0.93 0.93 143 ``` ] ??? --- #### The Precision-Recall curve SVC: the classifier on the Support Vector Machine algorithm ```python # [...] X_train, X_test, y_train, y_test = train_test_split(X, y) svc = SVC.fit(X_train, y_train) precision, recall, thresholds = precision_recall_curve(y_test, svc.decision_function(X_test)) ``` (some details were omitted) ??? --- #### The Precision-Recall curve of an SVC classifier .center[ 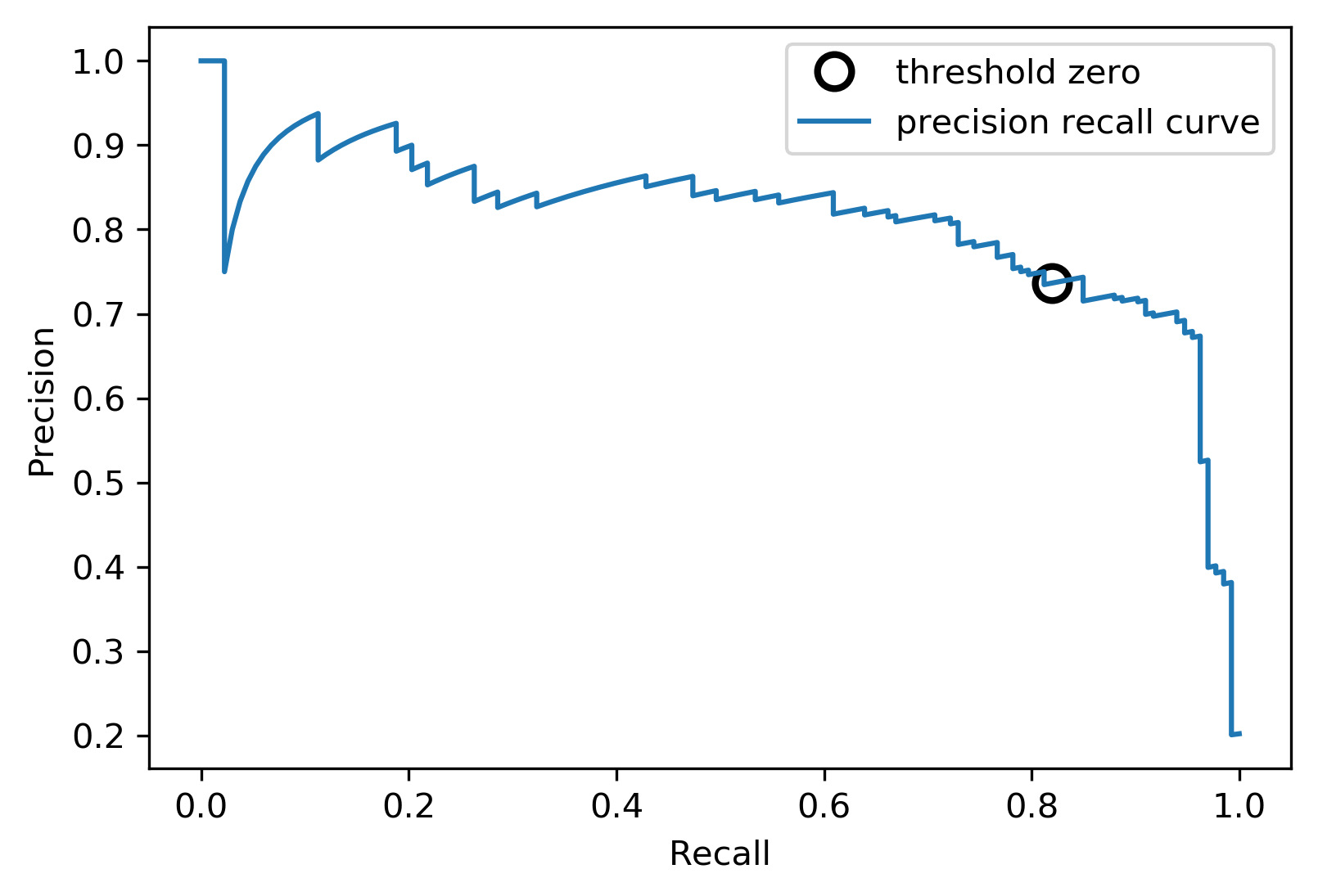 ] ??? --- #### Comparing RF and SVC .center[ 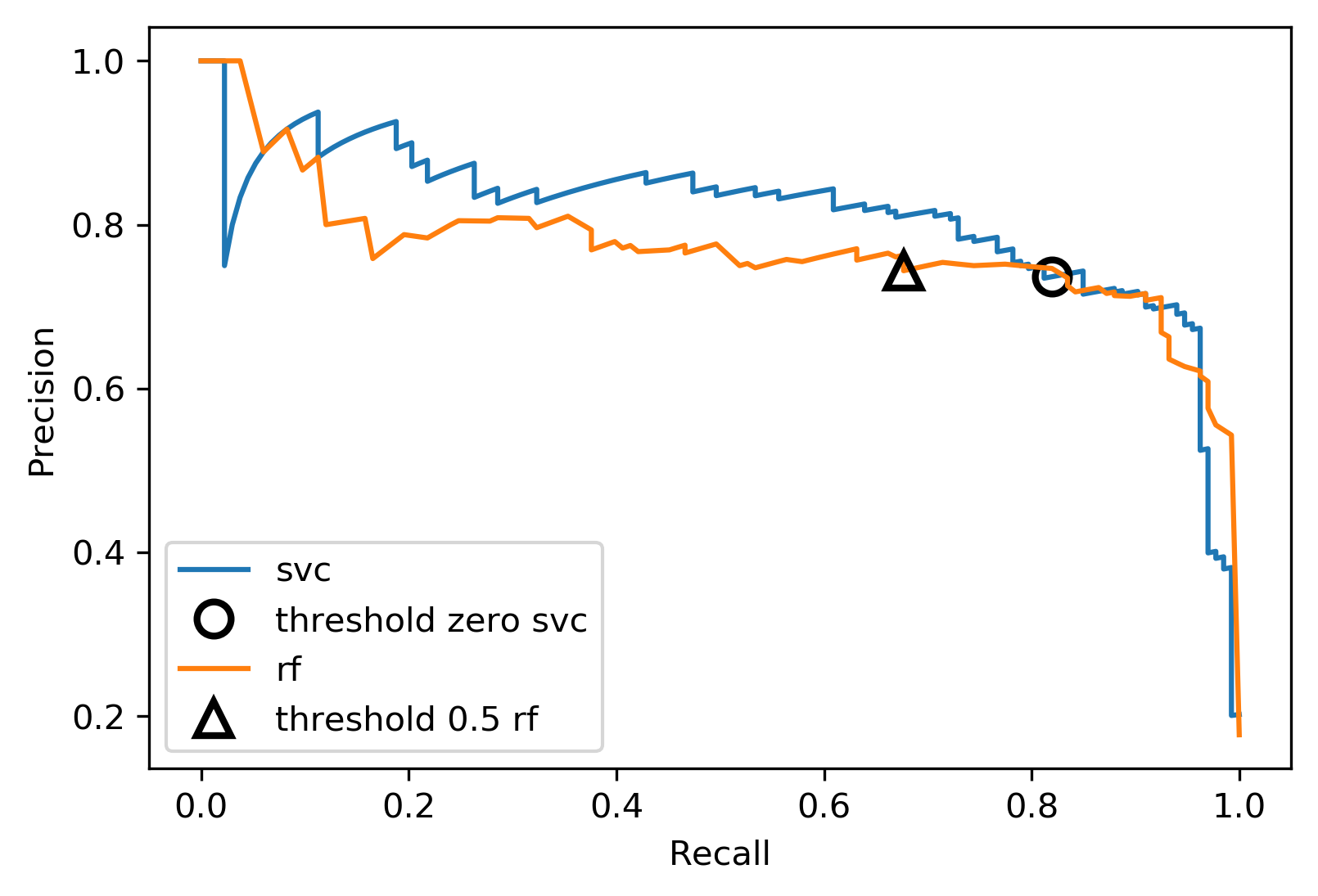 ] ??? --- #### Alternative: Average Precision .center[ 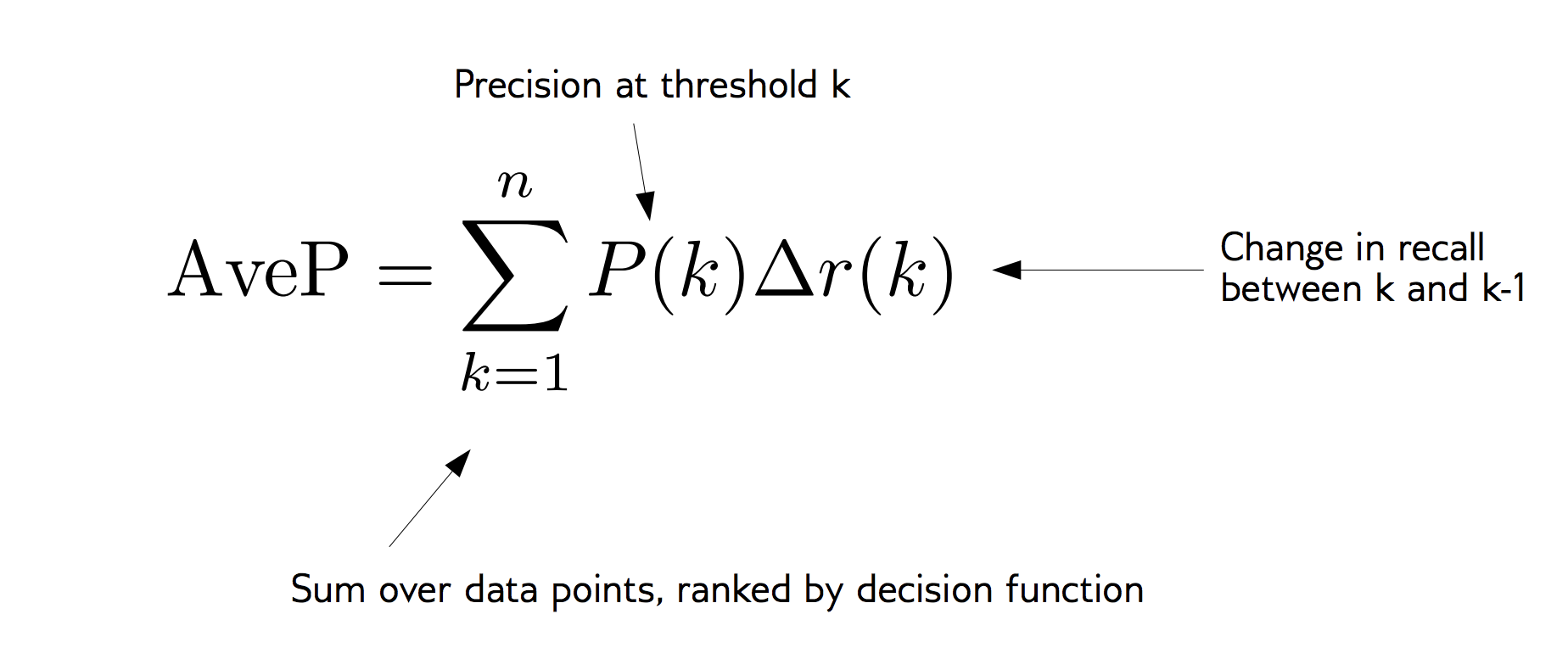 ] ??? Related to area under the precision-recall curve (with step interpolation) --- #### F1 vs. average Precision .smaller[ ```python from sklearn.metrics import f1_score print("f1_score of random forest: {:.3f}".format(f1_score(y_test, rf.predict(X_test)))) print("f1_score of svc: {:.3f}".format(f1_score(y_test, svc.predict(X_test)))) ``` ``` f1_score of random forest: 0.709 f1_score of svc: 0.715 ``` ```python from sklearn.metrics import average_precision_score ap_rf = average_precision_score(y_test, rf.predict_proba(X_test)[:, 1]) ap_svc = average_precision_score(y_test, svc.decision_function(X_test)) print("Average precision of random forest: {:.3f}".format(ap_rf)) print("Average precision of svc: {:.3f}".format(ap_svc)) ``` ``` Average precision of random forest: 0.682 Average precision of svc: 0.693 ``` ] ??? AP only considers ranking! --- #### The ROC Curve .center[  ] `$$ \text{FPR} = \frac{\text{FP}}{\text{FP}+\text{TN}}$$` `$$ \text{TPR} = \frac{\text{TP}}{\text{TP}+\text{FN}} = \text{recall}$$` ??? --- .center[ 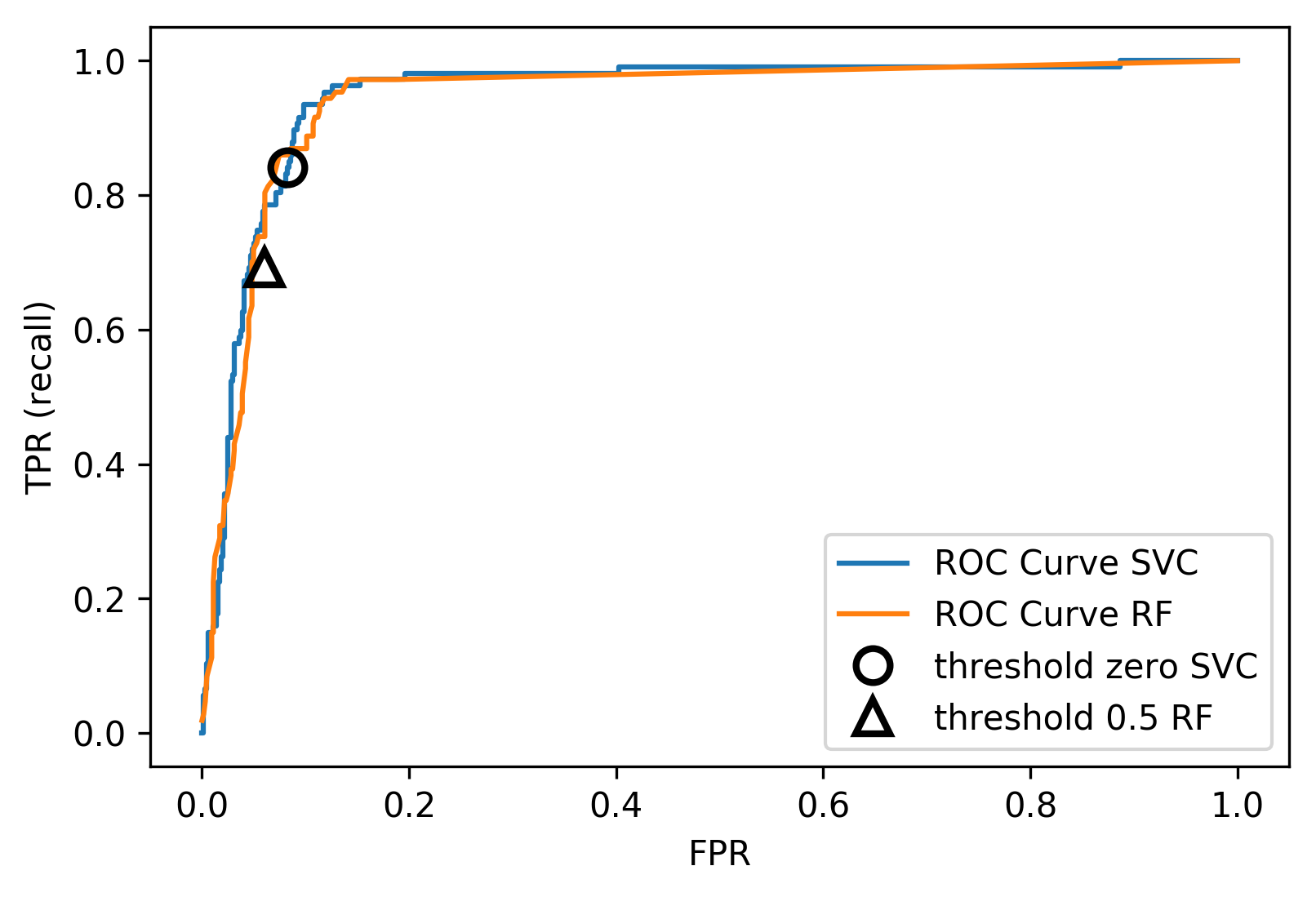 ] ??? --- #### The ROC AUC - Area under ROC Curve - Always .5 for random/constant prediction .tiny-code[ ```python from sklearn.metrics import roc_auc_score rf_auc = roc_auc_score(y_test, rf.predict_proba(X_test)[:,1]) svc_auc = roc_auc_score(y_test, svc.decision_function(X_test)) print("AUC for random forest: {:, .3f}".format(rf_auc)) print("AUC for SVC: {:, .3f}".format(svc_auc)) ``` ``` AUC for random forest: 0.937 AUC for SVC: 0.916 ``` ] Backgorund reading: *The Relationship Between Precision-Recall and ROC Curves* <br /> https://www.biostat.wisc.edu/~page/rocpr.pdf ??? --- #### Summary of metrics for binary classification Threshold-based: - precision, recall, - accuracy - `$F_\alpha$` Ranking-based: - average precision - ROC AUC add log-loss? ??? --- class: centre,middle ## Non-binary (Multi-class) classification ??? --- class:split-40 #### Confusion Matrix .left-column[ .tiny-code[ ```python from sklearn.datasets import load_digits from sklearn.metrics import accuracy_score digits = load_digits() # data is between 0 and 16 X_train, X_test, y_train, y_test = train_test_split(digits.data / 16., digits.target, random_state=0) lr = LogisticRegression().fit(X_train, y_train) pred = lr.predict(X_test) print("Accuracy: {:.3f}".format(accuracy_score(y_test, pred))) print("Confusion matrix:") print(confusion_matrix(y_test, pred)) ``` ] ] --- .tiny-code[ ```python [...] print("Accuracy: {:.3f}".format(accuracy_score(y_test, pred))) print("Confusion matrix:") print(confusion_matrix(y_test, pred)) ``` ``` Accuracy: 0.964 Confusion matrix: [[37 0 0 0 0 0 0 0 0 0] [ 0 41 0 0 0 0 1 0 1 0] [ 0 0 44 0 0 0 0 0 0 0] [ 0 0 0 43 0 0 0 0 1 1] [ 0 0 0 0 37 0 0 1 0 0] [ 0 0 0 0 0 47 0 0 0 1] [ 0 1 0 0 0 0 51 0 0 0] [ 0 0 0 0 1 0 0 47 0 0] [ 0 3 1 0 0 1 0 0 43 0] [ 0 0 0 0 0 2 0 0 1 44]] ``` ] ] --- .tiny-code[ ```python print(classification_report(y_test, pred)) ``` ``` precision recall f1-score support 0 1.00 1.00 1.00 37 1 0.91 0.95 0.93 43 2 0.98 1.00 0.99 44 3 1.00 0.96 0.98 45 4 0.97 0.97 0.97 38 5 0.94 0.98 0.96 48 6 0.98 0.98 0.98 52 7 0.98 0.98 0.98 48 8 0.93 0.90 0.91 48 9 0.96 0.94 0.95 47 avg / total 0.96 0.96 0.96 450 ``` ] ??? --- #### Averaging strategies - "macro", "weighted", "micro" (multi-label), "samples" (multi-label) `$$\text{macro: }\frac{1}{\left|L\right|} \sum_{l \in L} R(y_l, \hat{y}_l)$$` `$$\text{weighted: } \frac{1}{n} \sum_{l \in L} n_l R(y_l, \hat{y}_l)$$` .smaller[ ```python print("Micro average: ", recall_score(y_test, pred, average="weighted")) print("Macro average: ", recall_score(y_test, pred, average="macro")) ``` ``` Micro average: 0.964 Macro average: 0.964 ``` ] ??? micro vs macro same for other metrics. --- #### Multi-class ROC AUC - Hand & Till, 2001, one vs one `$$ \frac{1}{c(c-1)}\sum_{j=1}^{c}\sum_{k \neq j}^{c} AUC(j,k)$$` develop a classifier for each pair of possible labels; let them vote on each unseen input - Provost & Domingo, 2000, one vs rest `$$ \frac{1}{c}\sum_{j=1}^{c}p(j) AUC(j,\text{rest}_j)$$` Develop a classifier for each possible label Apply all of them to an unseen input and that give the label of the corresponding classifier that reports the highest *confidence* score --- #### observations: - Accuracy rarely what you want - Emphasis on recall or precision? - Problems are rarely balanced --- #### Available Scikit-learn scoring functions ```python from sklearn.metrics.scorer import SCORERS print("\n".join(sorted(SCORERS.keys()))) ``` .smaller[ ``` accuracy log_loss precision_micro adjusted_mutual_info_score mean_absolute_error precision_samples adjusted_rand_score mean_squared_error precision_weighted average_precision median_absolute_error r2 completeness_score mutual_info_score recall explained_variance neg_log_loss recall_macro f1 neg_mean_absolute_error recall_micro f1_macro neg_mean_squared_error recall_samples f1_micro neg_mean_squared_log_error recall_weighted f1_samples neg_median_absolute_error roc_auc f1_weighted normalized_mutual_info_score v_measure_score fowlkes_mallows_score precision homogeneity_score precision_macro ``` ]